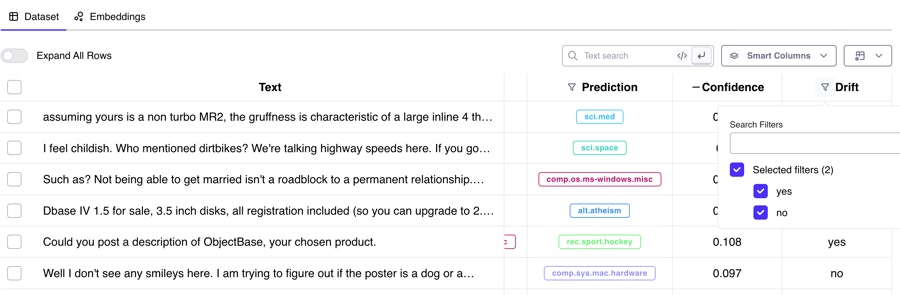
Text Classification
- How to find mislabeled samples?
- How to analyze misclassified samples?
- What is DEP and how to use it?
- How to inspect my model’s embeddings?
- How to best leverage Similarity Search?
Named Entity Recognition
- NER: What’s new?
-
How to identify spans that were hard to train on?
- Most Frequent High DEP words
- Span-level Embeddings
- What do the different Error Types mean?
Questions
- How do I install the Galileo Python client?
- I’m seeing errors importing dataquality in jupyter/google colab
- My run finished, but there’s no data in the console! What went wrong?
- Can I Log custom metadata to my dataset?
- How do I disable Galileo logging during model training?
- How do I load a Galileo exported file for re-training?
- How do I get my NER data into huggingface format?
- My spans JSON column for my NER data can’t be loaded with json.loads
- Galileo marked an incorrect span as a span shift error, but it looks like a wront tag error. What’s going on?
- What do you mean when you say the deployment logs are written to Google Cloud?
- Does Galileo store data in the cloud?
- Where are the client logs stored?
- Do you offer air-gapped deployments?
- How do I contact Galileo?
- How do I convert my vaex dataframe to pandas when using dq.metrics.get_dataframe?
- Importing dataquality throws a permissions error `PermissionError`
- vaex-core fails to build with Python 3.10 on MacOs Monterey
- Training a model is really slow. Can I make it go faster?
Q: How do I install the Galileo Python client?
Q: I’m seeing errors importing dataquality in Jupyter / Google Colab
Make sure you running at leastdataquality >= 0.8.6 The first thing to try in this case it to restart your kernel. Dataquality uses certain python packages that require your kernel to be restarted after installation. In Jupyter you can click “Kernel -> Restart”
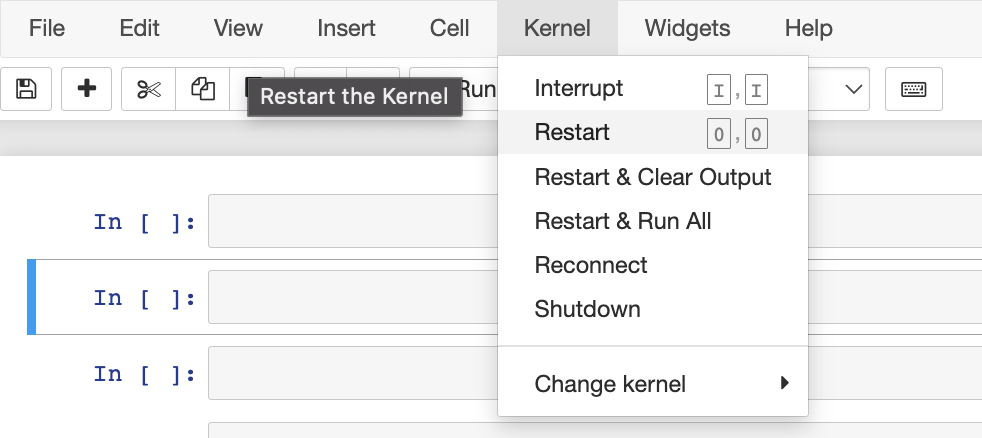
dataquality, there is a known bug when upgrading. Solution: pip uninstall -y vaex-core vaex-hdf5 && pip install --upgrade --force-reinstall dataquality “And then restart your jupyter/colab kernel
Q: My run finished, but there’s no data in the console! What went wrong?
Make sure you randq.finish() after the run.
t’s possible that:
- your run hasn’t finished processing
- you’ve logged some data incorrectly
- you may have found a bug (congrats!
dq.wait_for_run() (you can optionally pass in the project and run name, or the most recent will be used)
This function will wait for your run to finish processing. If it’s completed, check the console again by refreshing.
If that shows an exception, your run failed to be processed. You can see the logs from your model training by running dq.get_dq_log_file() which will download and return the path to your logfile. That may indicate the issue. Feel free to reach out to us for more help!
Q: Can I log custom metadata to my dataset?
Yes (glad you asked)! You can attach any metadata fields you’d like to your original dataset, as long as they are primitive datatypes (numbers and strings). In all available logging functions for input data, you can attach custom metadata: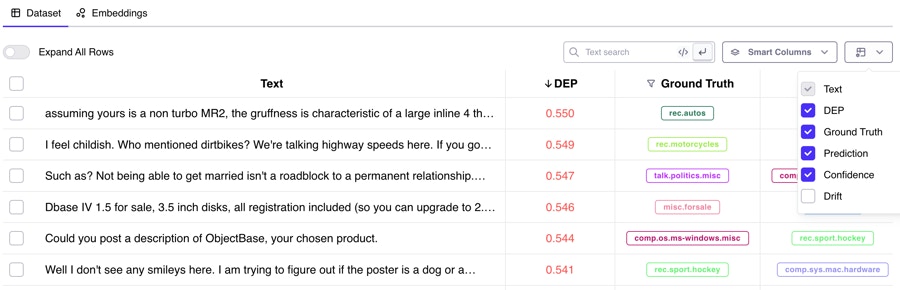
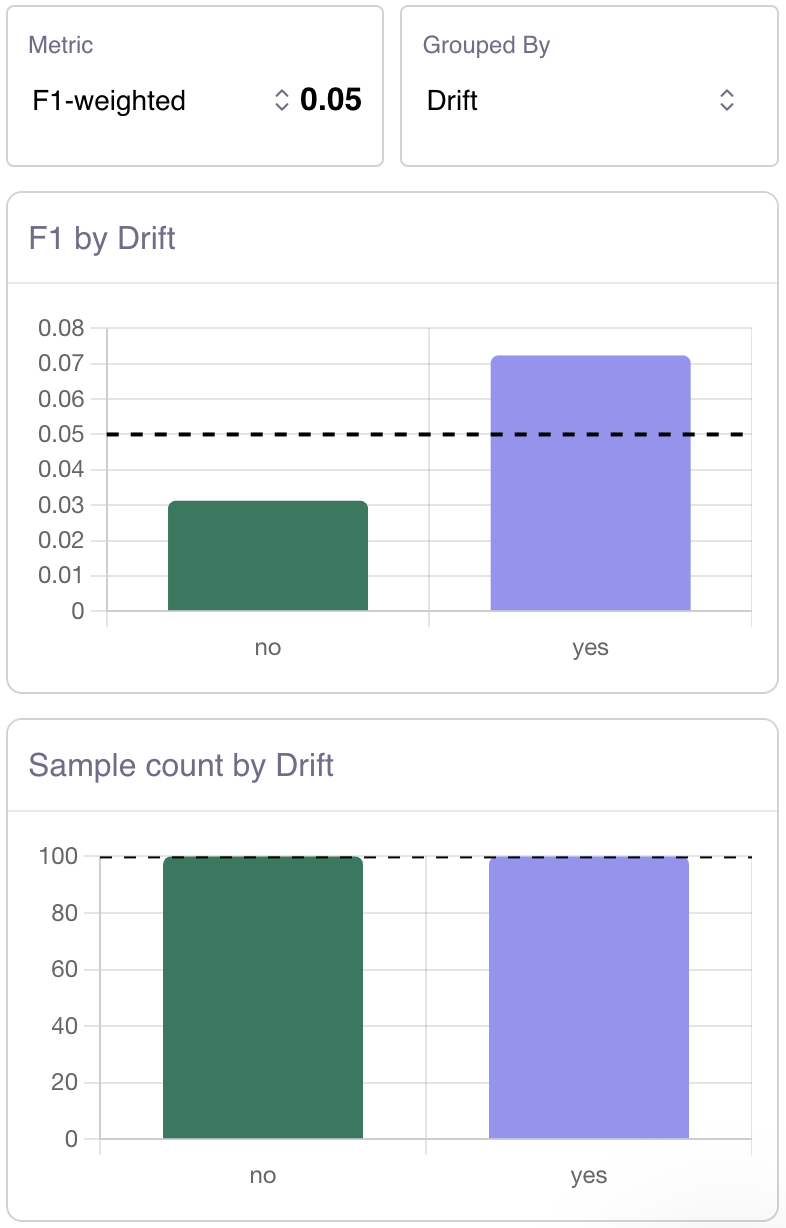
Q: How do I disable Galileo logging during model training?
See Disabling Galileo
Q: How do I load a Galileo exported file for re-training?
Q: How do I get my NER data into huggingface format?
Q: My spans JSON column for my NER data can’t be loaded with json.loads
If you’re seeing an error similar to: JSONDecodeError: Expecting ',' delimiter: line 1 column 84 (char 83) It’s likely the case that you have some data in your text field that is not valid json (extra quotes " or '). Unfortunately, we cannot modify the content of your span text, but we can strip out the text field with some regex. Given a pandas dataframe df with column spans (from a Galileo export) you can replace df["spans"] = df.apply(json.loads) with (make sure to import re) df["spans"] = df.apply(lambda row: json.loads(re.sub(r","text".}", "}", row)))
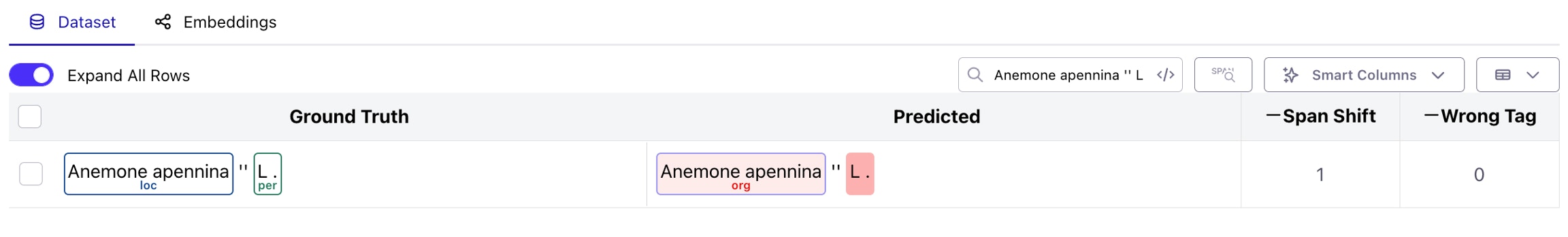
Q: Galileo marked an incorrect span as a span shift error, but it looks like a wrong tag error. What’s going on?
Great observation! Let’s take a real example below, from the WikiNER IT dataset. As you can see, theAnemone apennina clearly looks like a wrong tag error (correct span boundaries, incorrect class prediction), but is marked as a span shift.
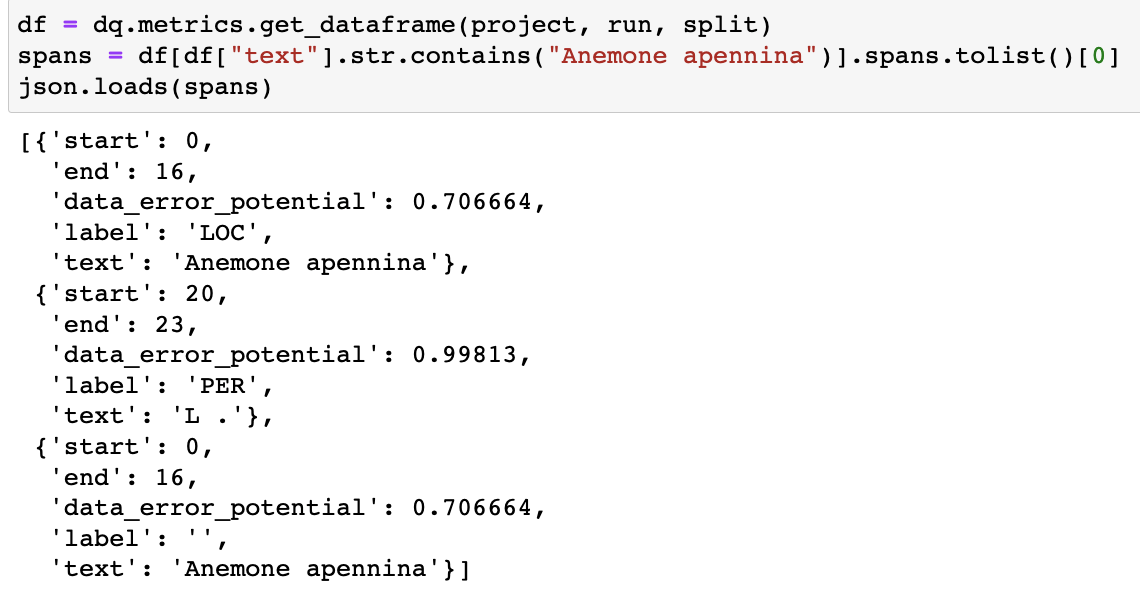
dq.metrics.get_dataframe. We can see that there are 2 spans with identical character boundaries, one with a label and one without (which is the prediction span).
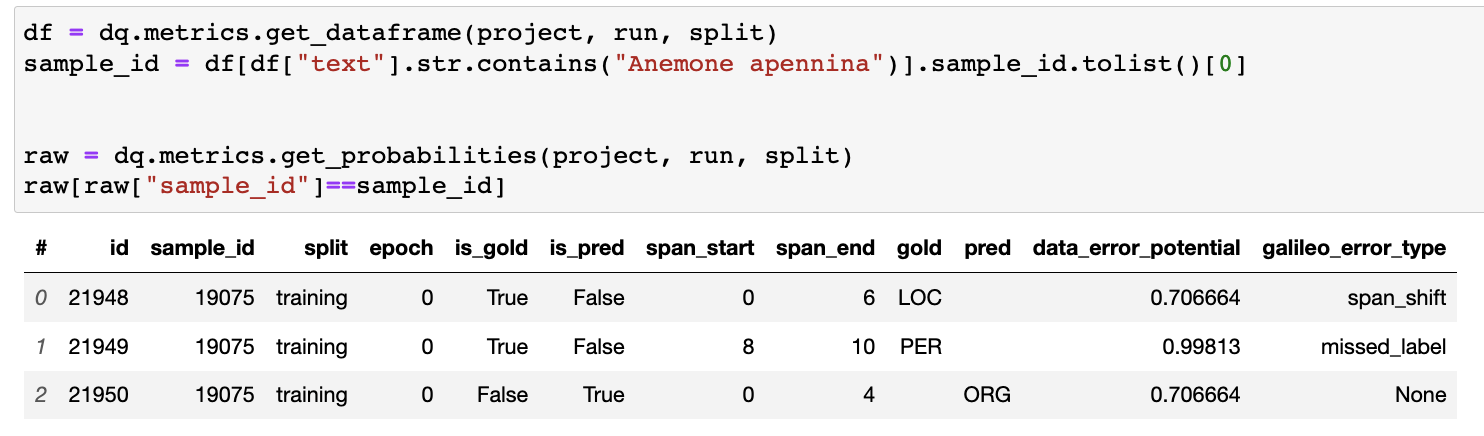
dq.metrics by looking at the raw data logged to Galileo. As we can see, at the token level, the span start and end indices do not align, and in fact overlap (ids 21948 and 21950), which is the reason for the span_shift error
Q: What do you mean when you say the deployment logs are written to Google Cloud?
We manage deployments and updates to the versions of services running in your cluster via Github Actions. Each deployment/update produces logs that go into a bucket on Galileo’s cloud (GCP). During our private deployment process **** (for Enterprise users), we allow customers to provide us with their emails, so they can have access to these deployment logs.Q: Where are the client logs stored?
The client logs are stored in the home (~) folder of the machine where the training occurs.Q: Does Galileo store data in the cloud?
For Enterprise Users, data does not leave the customer VPC/Data Center. For users of the Free version of our product, we store data and model outputs in secured servers in the cloud. We pride ourselves in taking data security very seriously.Q: Do you offer air-gapped deployments?
Yes, we do! Contact us to learn more.Q: How do I contact Galileo?
You can write us at team[at]rungalileo.ioQ: How do I convert my vaex dataframe to a pandas DataFrame when using the dq.metrics.get_dataframe
Simply add dq.metrics.get_dataframe(...).to_pandas_df()
Importing dataquality throws a permissions error **PermissionError**
Galileo creates a folder in your system’s HOME directory. If you are seeing a PermissionsError it means that your system does not have access to your current HOME directory. This may happen in an automated CI system like AWS Glue. To overcome this, simply change your HOME python Environment Variable to somewhere accessible. For example, the current directory you are in
HOME directory. Because of that, if you run a new python script in this environment again, you will need to set the HOME variable in each new runtime.
Q: vaex-core fails to build with Python 3.10 on MacOs Monterey
When installing dataquality with python 3.10 on MacOS Monterey you might encounter an issue when building vaex-core binaries. To fix any issues that come up, please follow the instructions in the failure output which may include runningxcodebuild -runFirstLaunch and also allowing for any clang permission requests that pop up.
Q: Training a model is really slow. Can I make it go faster?
For larger datasets you can speed up model training by running CUDA. Note: You must be running CUDA 11.X for this functionality to work. Cuda’s CUML libraries require CUDA 11.X to work properly. You can check your CUDA version by runningnvcc -V. Do not run nvidia-smi, that does not give you the true CUDA version. To learn more about this installation or to do it manually, see the installation guide.
If you are training on datasets in the millions, and noticing that the Galileo processing is slowing down at the “Dimensionality Reduction” stage, you can optionally run those steps on the GPU/TPU that you are training your model with.
In order to leverage this feature, simply install dataquality with the [cuda] extra.
extra-index-url to the install, because the extra required packages are hosted by Nvidia, and exist on Nvidia’s personal pypi repository, not the standard pypi repository.
After running that installation, dataquality will automatically pick up on the available libraries, and leverage your GPU/TPU to apply the dimensionality reduction.
Please validate that the installation ran correctly by running import cuml in your environment. This must complete successfully.
To manually install these packages (at your own risk), you can run